The Goal
Fundamental to all biological systems are proteins as well as machinery for synthesizing, folding, translocating, and degrading each component of the proteome. Collectively, these processes comprise proteostasis, which is essential for maintaining the proteome in a healthy state. The components that maintain proteostasis are together known as the proteostasis network. Various diseases are characterized by failures of proteostasis, including the aging-related neurodegenerative diseases, metabolic diseases, muscle wasting diseases, and cancers. The widespread interest in these diseases suggests that having a comprehensive description of the proteostasis network would be invaluable. With a curated and annotated list of all proteostasis network genes in hand, research groups will be able to efficiently evaluate proteostasis variations in genome-wide data sets. By gaining broader and systematic insights into the relationship between proteostasis and disease states, identifying critical limiting targets for therapeutic interventions should be closer at hand.
The Proteostasis Network Team
Practically speaking, individual laboratories maintain expertise in individual branches of the proteostasis network, rather than the whole of it. Over the course of more than two years, members of the Finkbeiner (University of California San Francisco and Gladstone Institute), Finley (Harvard Medical School), Frydman (Stanford University), Gestwicki (UCSF), Kelly (Scripps Research), and Morimoto (Northwestern University) laboratories have been working together to assemble the annotated proteostasis network, drawing on subject area expertise as well as a shared vision of how the proteostasis network should be defined, described, and presented. They have now been joined by members of the Cuervo (Einstein Medical) and Prado (ISPA) laboratories.

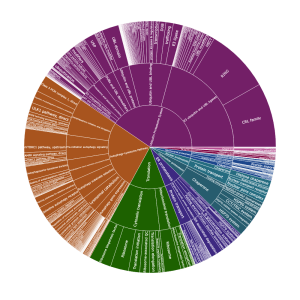
Figure 1- Branches of the Human Proteostasis Network
Scope of the Proteostasis Network
There are nine main divisions of the proteostasis network: mitochondrial proteostasis, ER proteostasis, cytonuclear proteostasis, nuclear proteostasis, cytosolic translation, proteostasis regulation, extracellular proteostasis, the autophagy lysosome pathway (ALP), and the ubiquitin proteasome system (UPS). Note that the term cytonuclear refers to components that support proteostasis in both the cytosol and the nucleus, whereas nuclear refers to components that primarily support nuclear proteostasis specifically. Also note that components of proteostasis regulation may act at either the transcriptional or translational level. Genes may be listed in more than one branch, and the nonredundant gene list currently comprises almost 3000 members while the total number of annotations is close to 3,800.
Methodology for Enumerating the Proteostasis Network
We used entity-based and domain-based criteria to determine whether a given component should be included in the proteostasis network. Entity-based refers to inclusion of a gene product (a protein or non-coding RNA) based on positive biochemical, cell biological, or genetic evidence in the literature that it functions in proteostasis. Domain-based refers to inclusion based on a protein containing a structural domain that is strongly associated with proteostasis; for example, the J-domain, which characterizes cochaperones in the HSP70 system. Preliminary lists of proteostasis network components were generated based on functional databases (like KEGG and Gene Ontology), reviews, other literature on the relevant systems, and conversations with subject matter experts within and outside of the Consortium. These preliminary lists were then vetted gene by gene by members of the Consortium. For borderline cases, rationales for inclusion or exclusion were presented by Proteostasis Consortium members with subject area expertise and a decision was made based on the consensus of the group.
Taxonomic Annotation of the Proteostasis Network

We sought an annotation system that would convey at a glance and for any component both its specific function in proteostasis and how it relates to the rest of the proteostasis network. Thus, we developed a simple taxonomic scheme consisting of just five levels: Branch, Class, Group, Type, and Subtype. The broadest category, Branch, refers to a component’s localization or membership in an overarching pathway. The nine branches are listed above and are arrayed around the cell in Figure 1. Class refers to a component’s function in proteostasis (e.g., chaperones, protein transport), while Group, Type, and Subtype provide increasingly specific descriptors of proteostasis functions within a Class. Our taxonomy as applied to the BAG domain family proteins is shown at right. Our goal was to use only as many descriptors as are minimally necessary to give a basic understanding of a component’s role in proteostasis. Thus, not every component has Type or Subtype annotations. Also, some components have multiple roles in the proteostasis network. These are given multiple entries in our list to reflect their separate roles. The current Class-level breakdowns of all the Branches are shown in Figure 2 below, where the nine Branches are shown in central boxes and their Classes are in the boxes to the left or right. The numbers in gray boxes correspond to the numbers of unique components in each Class.
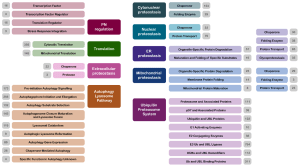
Figure 2- Branches and Classes of the Human Proteostasis Network
Interactive Visualization of the Proteostasis Network Taxonomy
Users can explore the proteostasis network’s taxonomy in this interactive sunburst chart. Starting from the broadest Branch level, it narrows down through Class, Group, Type, and Subtype, each providing finer detail about components’ roles in proteostasis. The chart’s segments, colored by category and proportioned by number, offer a glance into the network’s complexity, with additional information on hover.
Downloading the Annotated Proteostasis Network
Annotation of the human proteostasis network branches was originally presented in three tranches, as described below. Here we present a second version of the Human Proteostasis Network Annotation. The scope of the original annotation has increased somewhat and includes both new genes and new annotations. The annotation now includes four indexing values: Gene Symbols, Gene IDs, Uniprot IDs, and Ensembl IDs, allowing easy engagement with all bioinformatic tools.
The file below presents the network in a few formats, and reading the ReadMe tab is highly recommended.
Use of the resources presented on this page should be acknowledged by citing this website or by citing the bioRχiv papers listed below.
Development of the ALP branch and the branches concerning chaperones, the protein synthesis machinery, trafficking systems, and organelle-specific degradation systems was led by Evan Powers. Please direct questions or comments about these branches to him via PNAnnotation@gmail.com. Work on the UPS was led by Suzanne Elsasser. Please direct questions or comments about the UPS to Suzanne Elsasser (suzanne_elsasser@hms.harvard.edu) and Daniel Finley (daniel_finley@hms.harvard.edu).
The PN team is indebted to external collaborators for their assistance and expertise. Many collaborators immeasurably enhanced the development of proteostasis network branches: for the UPS, Kay Hofmann (University of Cologne) and Alfred Vertegaal (Leiden University Medical Center); for the ALP, Ralph A. Nixon (NYU Langone Health); for the ER and mitochondria, R. Luke Wiseman (Scripps Research); and for extracellular proteostasis, Joel N. Buxbaum (Scripps Research).
View the bioRχiv papers
Two bioRχiv papers have been written to chronicle this work, covering all branches except the UPS, which will be forthcoming.
A Comprehensive Enumeration of the Human Proteostasis Network. 1. Components of Translation, Protein Folding, and Organelle-Specific Systems doi:https://doi.org/10.1101/2022.08.30.505920
Initial versions of the Human PN Annotation
The initial annotation of the Human Proteostasis Network was released as three tranches, the first two tranches corresponding to the first two bioRχiv papers. The file below contains those three tranches which appear in three separate tabs. This version of the Human PN Annotation has been superseded by version 2.0 (see above). The original files are provided as a reference particularly for those who have already developed analyses based on the original annotation.
Proteostasis Network 1.0 ~ 2022 and 2023 (1)
Page last updated: 04/23/2024