Core A | Core B | Core C | Core D
Core A: Administration
The Administrative Core at Northwestern is responsible for:
- Promoting a stimulating environment for the entire Proteostasis Consortium
- Supporting project-based collaborations across member laboratories
- Supporting diversity and outreach
Core Leader: Richard Morimoto, (Northwestern)
Business Administrator: Please contact Rebecca Phend (rebecca.phend@northwestern.edu)
Core B: Sensors and tools for proteostasis analysis
Core Co-Leaders: Judith Frydman (Stanford) and Steve Finkbeiner (Gladstone/UCSF)
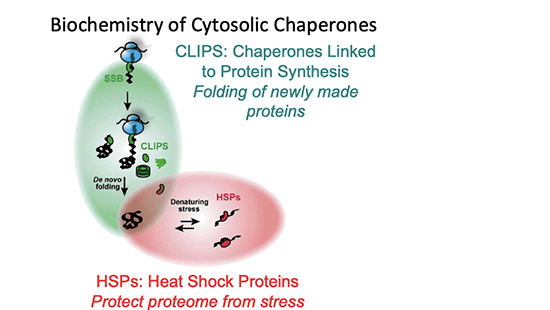 This Core will develop tools to define the Proteostasis Network (PN during aging and in the context of disease-associated aggregation-prone proteins and will provide technical support to Projects 1–4. These resources will test and validate compounds from Core D. Core B will be located at two sites in close geographical proximity. The Frydman group (Stanford) will build molecular sensors and assays to measure proteostasis changes during aging, starting with reporters developed for yeast chaperone machinery and the ubiquitin-proteasome system (UPS). The Finkbeiner group (Gladstone/UCSF) will build mammalian expression and analysis tools to assess protein aggregation and autophagy and help other Projects address specific questions for which their multiplexed longitudinal single-cell imaging platform is critical. By working hand-in-hand and testing the ability of the sensors to report on the proteostasis events used by all Projects, this Core will produce a set of assays for a comprehensive view of the PN in all cells. These data will be placed on the intranet managed by Core A and upon publication will be released to the entire community.
This Core will develop tools to define the Proteostasis Network (PN during aging and in the context of disease-associated aggregation-prone proteins and will provide technical support to Projects 1–4. These resources will test and validate compounds from Core D. Core B will be located at two sites in close geographical proximity. The Frydman group (Stanford) will build molecular sensors and assays to measure proteostasis changes during aging, starting with reporters developed for yeast chaperone machinery and the ubiquitin-proteasome system (UPS). The Finkbeiner group (Gladstone/UCSF) will build mammalian expression and analysis tools to assess protein aggregation and autophagy and help other Projects address specific questions for which their multiplexed longitudinal single-cell imaging platform is critical. By working hand-in-hand and testing the ability of the sensors to report on the proteostasis events used by all Projects, this Core will produce a set of assays for a comprehensive view of the PN in all cells. These data will be placed on the intranet managed by Core A and upon publication will be released to the entire community.
Specific Aim
- To develop Proteostasis Network reporters to quantify different arms of Proteostasis
By synthesizing knowledge of the PN into a set of validated fluorescence-based sensors and reporters, the core will produce an invaluable toolbox for the whole proteostasis field that will facilitate progress, not only in the labs associated with this PPG, but in other labs interested in linking proteostasis dysfunction with disease.- To develop and implement improved analysis modalities using quantitative image analyses and time-resolved image analyses.
- To develop and optimize a multiplexed orthogonal panel of reporters for temporal investigation and implement novel approaches, such as hyperspectral longitudinal single-cell analysis.
- To employ deep learning to analyze the data to more fully understand how proteostasis functions as a network in Alzheimer’s disease.
Core C: Phenotypic analysis and global proteostasis network assessment by TMT-MS3 proteomics
Core Leader: Dan Finley (Harvard)
Team Leader: Miguel Prado (ISPA Institute, Spain)
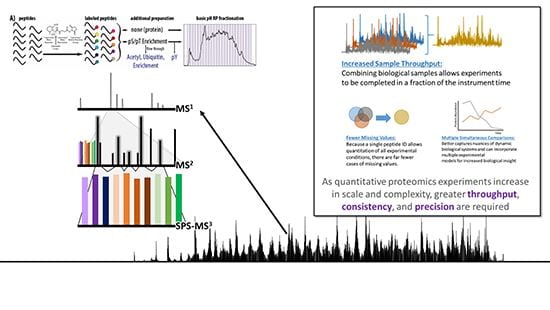 This Core will perform proteomic analyses of samples from all participating groups and will be a foundational approach for all Projects. This will also allow assessment of the effects of GRN deficiency on the levels of proteins that are autophagy substrates, including components of organelles, such as mitochondria, to assess the effects on the autophagy lysosomal pathway (ALP) more broadly on proteolysis. The TMT-MS3 experiments will provide a snapshot of the status of the proteostasis network (PN), with complete descriptions on the levels of hundreds of chaperones and components of the UPS and UPR. The Core will work with Project 2 to determine proteasome levels in transgenic mice that conditionally express the proteasome subunits Rpn6, Rpn11, or ß5 from both the 19-subunit regulatory particle and 28-subunit core particle of the proteasome that should elevate overall proteasome levels. Finally, Core C will work closely with Cores B and D to demonstrate the relationship between the properties of Proteostasis Sensors and their response to small molecule Proteostasis Regulators.
This Core will perform proteomic analyses of samples from all participating groups and will be a foundational approach for all Projects. This will also allow assessment of the effects of GRN deficiency on the levels of proteins that are autophagy substrates, including components of organelles, such as mitochondria, to assess the effects on the autophagy lysosomal pathway (ALP) more broadly on proteolysis. The TMT-MS3 experiments will provide a snapshot of the status of the proteostasis network (PN), with complete descriptions on the levels of hundreds of chaperones and components of the UPS and UPR. The Core will work with Project 2 to determine proteasome levels in transgenic mice that conditionally express the proteasome subunits Rpn6, Rpn11, or ß5 from both the 19-subunit regulatory particle and 28-subunit core particle of the proteasome that should elevate overall proteasome levels. Finally, Core C will work closely with Cores B and D to demonstrate the relationship between the properties of Proteostasis Sensors and their response to small molecule Proteostasis Regulators.
Specific Aim
- Proteostasis Proteomics Core C will provide and develop novel data and tools for PN function and client interactions.
- This core will conduct global proteomics experiments under the leadership of Steve Gygi and Daniel Finley at Harvard Medical School to provide quantitative readouts of protein levels of 10 samples at once, using the TMT-MS3 method of global proteomics developed by the Gygi group. A single run of 10 samples provides detailed quantitative data on 10 experimental proteomes.
- This comprehensive view of perturbations under defined experimental conditions will yield a wealth of information, including the levels of over 1000 components of the PN, such as proteasomes and chaperones, the state of thousands of substrates of these enzymes, and the occupancy of ~30,000 protein modification sites. Importantly, each TMT-MS3 run will provide a fingerprint of the state of the PN at a given time and condition.
Core D: Proteostasis regulator small molecule pharmacology core
Core Leader: Jeff Kelly (Scripps)
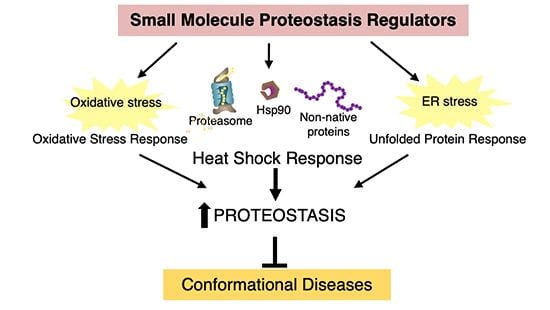 This Core together with Projects 1-4 and Cores B and C, is to test the hypothesis that it is possible to partially reverse aging-dependent deficiencies in proteostasis network capacity (including those leading to pathology) by pharmacologic regulation of the proteostasis network employing small molecule proteostasis regulators (PR).
This Core together with Projects 1-4 and Cores B and C, is to test the hypothesis that it is possible to partially reverse aging-dependent deficiencies in proteostasis network capacity (including those leading to pathology) by pharmacologic regulation of the proteostasis network employing small molecule proteostasis regulators (PR).
Since there are > 2000 targets within the proteostasis network (PN), the efforts will be focused on validating stress-responsive signaling pathway activators, and activators of the proteasome and autophagy degradation pathways. This will be accomplished by developing a technology platform to characterize the pharmacodynamics, selectivity, and mechanism of action of small molecule proteostasis regulators that function through activation of stress-responsive signaling pathways. The synthetic capabilities of the Kelly group will synthesize published candidate small molecule PRs that activate stress-responsive signaling pathways. The Core will characterize the pharmacokinetics (PK; the study of what the organism does to a drug (metabolism)) and pharmacodynamics (PD; the study of what a drug does to the organism) of candidate PRs in human cell culture, yeast cells, C. elegans, and in mice, to discern which compounds have the potential to pharmacologically regulate the PN in the organisms employed in this PPG, and to deliver validated PRs to PIs of the projects as well as labs working on complementary aging-associated diseases.
Specific Aims
- To test the hypothesis that it is possible to reverse aging-dependent deficiencies in PN capacity leading to neurodegeneration by pharmacologic regulation of the PN.
- To emphasize heat shock response activators that regulate cytosolic PN capacity, unfolded protein response activators that regulate secretory pathway capacity, the antioxidant stress-responsive pathway, proteasome activators and modulators of the autophagy lysosomal pathway.
- To employ our technology platform to validate PD, selectivity and MOA of PRs in cells and tissues of multiple organisms using reporters of stress-responsive signaling pathways, targeted RNAseq, LC-MS proteomics and bioinformatics to validate the PRs.
Stress-responsive signaling pathways generate active TFs that provide a greater opportunity for an effective biological response to correct proteostasis deficiencies, wherein all the components of interacting and competing PN pathways in a given subcellular compartment are up-regulated in the appropriate stoichiometry. These stress-responsive signaling pathways lead to powerful emergent functions that are only partially understood.
- To assess PK in multiple organisms, to establish reasonable dosing regimens. PK will be assessed by LC-MS.
- To establish the utility of PRs to enhance the degradation of proteins, lipids and organelles, either through activation of autophagy or the ubiquitin proteasome system.
To generate PK and PD for PRs reported by others that activate the autophagy lysosomal pathway through an m-TOR independent mechanism and for PRs to activate the ubiquitin proteasome system.

